SyBiOnt: The Synthetic Biology Ontology
Göksel Mısırlı, Jennifer Hallinan, Matthew Pocock, Phillip Lord, James Alastair McLaughlin, Herbert Sauro, and Anil Wipat. (2016), Data Integration and Mining for Synthetic Biology Design, ACS Synthetic Biology, 5(10), 1086-1097
SyBiOnt is an application ontology for synthetic biology. We developed this ontology to represent the richset of biological knowledge about biological components and their relationships. We demonstrated the use of this ontology to create the SyBiOntKB knowledge base, incorporating and building upon existing life sciences ontologies and standards. The reasoning capabilities of ontologies were then applied to automate the mining of biological parts from this knowledge base. This approach is be useful to speed up synthetic biology design and ultimately help facilitate the automation of the biological engineering life cycle.
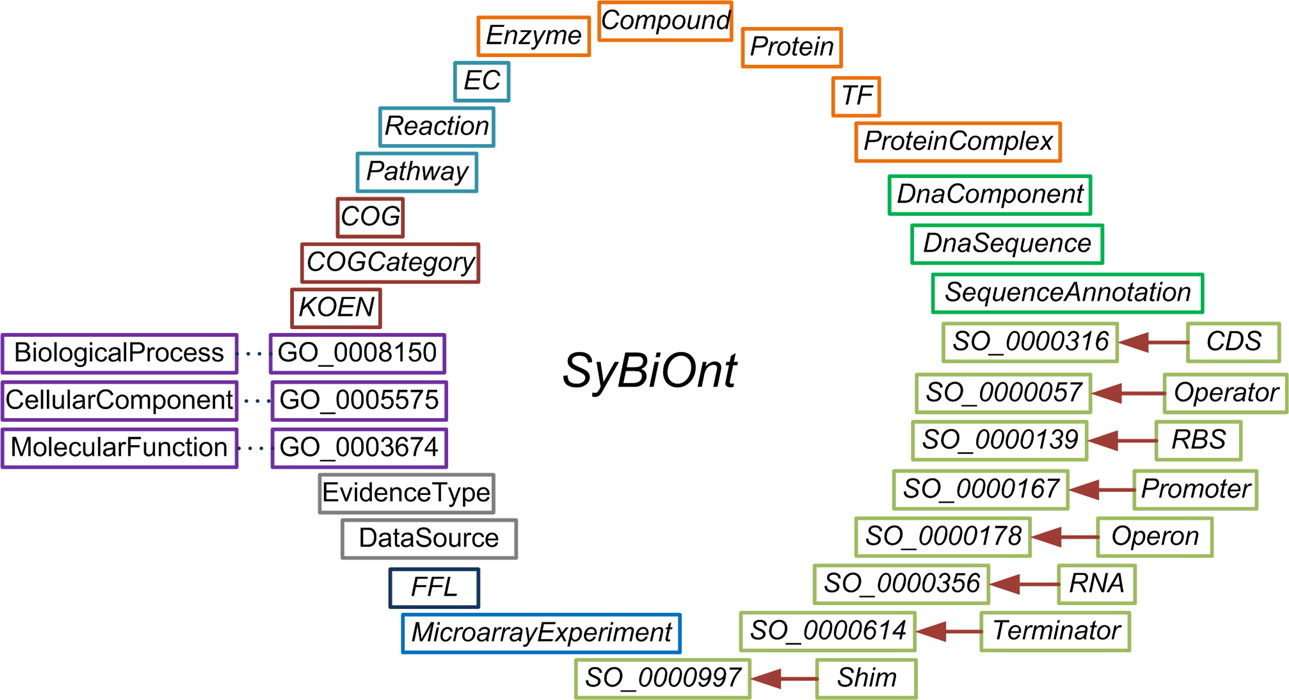
The basic biological parts used in the bottom-up design of synthetic systems include genetic features such as promoters, coding sequences (CDSs), ribosome binding sites (RBSs), terminators, and operators.The relationships among these parts and the gene products they encode, such as proteins, RNAs, transcription factors (TFs), and enzymes, need to be captured in order to design genetic circuits. Moreover, the incorporation of additional information about biological pathways and gene function is necessary to identify appropriate biological parts. Our goal when creating SyBiOnt was to allow a data definition framework to formalize the representation of the information that describes these parts and the relationships among them. SyBiOnt was designed to allow the incorporation of further information in the form of annotations that add extra, useful knowledge such as gene function. The ontology was developed using OWL semantics. The rich expressivity of OWL enables the construction of complex computational queries and automated reasoning across the integrated data.
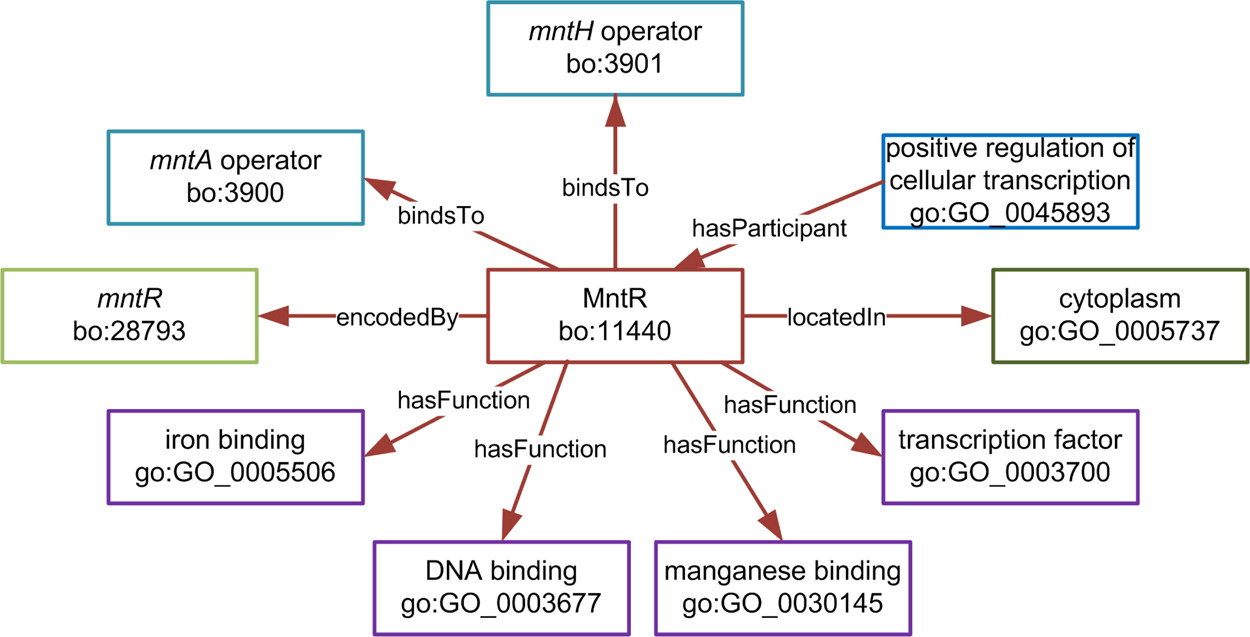
As an example of the use of the SyBiOnt ontology, we used the formal data definition framework provided to develop a knowledge base, termed SyBiOntKB, to capture major aspects of the cell biology of Bacillus subtilis in a computationally amenable form. The data to populate this knowledge base were sourced from the previously integrated BacillOndex data set, which includes information from BacilluScope, DBTBS, the Kyoto Encyclopedia of Genes and Genomes (KEGG), KEGG Expression, STRING, GO, and GO annotations. An example biological network, representing the relationships of the MntR transcription factor and how it relates to different biological concepts can be seen in the figure below.
SyBiOnt can be used to answer certain types of questions in a richer fashion than a conventional relational database. As an example, we showed how automated reasoning over this ontology could be used to identify parts and devices that could be used in synthetic designs. Particularly, we focused on the automated identification of promoters that could be used as logic gates (such as inducible or repressible), the building blocks of many synthetic biology designs. We then demonstrated the mining of CDS parts based on the molecular functions of their encoded products. In principle, the textual descriptions of classes from the ontology could be read by eye and used by humans to make assertions manually, but the use of automated reasoning vastly speeds up the process. The example below shows an ontological query, in the form of a class definition. When a reasoner is executed using the SyBiOnt, resources from the knowledge base matching this definition are classified as inducible promoters.
